
Coral Bleaching
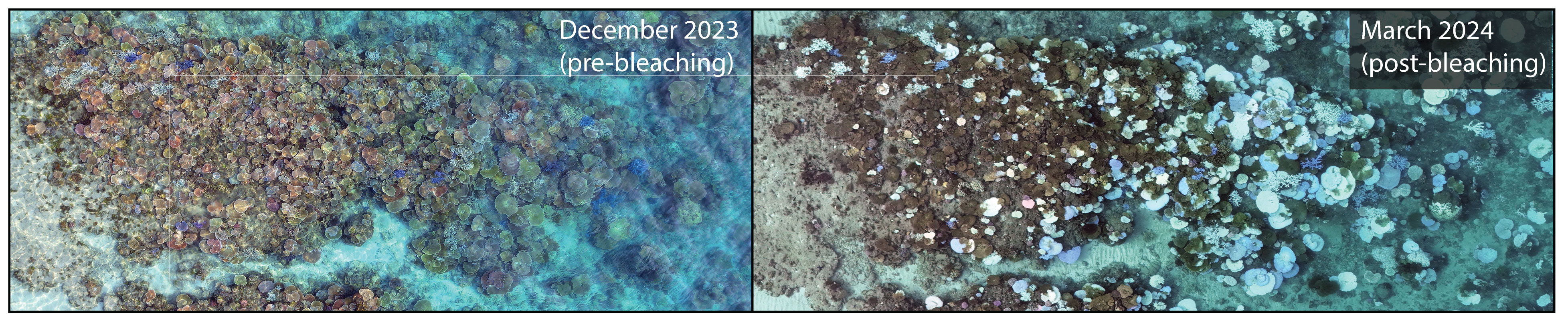
2024 “no bleaching” scenario
Code
library(tidyverse)
library(sf)
library(reefspawn)
seedval = 321
coral_size_2024 <- simulate_coralsize(coralsize |> filter(year==2011),
distribution = "rlnorm",
ndraws = 2000,
seed = seedval)
sf_plot <- setplot(20,10) # default EPSG:3857
coralsizepredictions <- posterior_coral_predict(brm = brm_sizedistribution,
newdata = coral_size_2024,
seed = seedval)
communities <- simulate_community(setplot = sf_plot,
coralcover = 40,
size = coral_size_2024,
seed = seedval)
populations <- simulate_populations(setplot = sf_plot,
size = coral_size_2024,
community = communities,
return="sf",
seed = seedval)
reproductive_populations <- simulate_spawning(populations = populations,
setplot = sf_plot,
seed = 123)
map_populations(setplot = sf_plot,
populations = reproductive_populations, # or reproductive output, same sf population
interactive = TRUE)2024 “bleaching” scenario
Code
reproductive_populations_postbleaching <- simulate_bleaching(populations = reproductive_populations)
map_populations(setplot = sf_plot,
populations = reproductive_populations_postbleaching,
interactive = TRUE)Mortality estimates
Code
library(DT)
# colony level info:
colony_output <- reproductive_populations_postbleaching |>
as.data.frame() |>
group_by(status, species) |>
summarise(n = n(), .groups = "drop") |>
pivot_wider(names_from = status, values_from = n, values_fill = 0) |>
mutate(
prebleaching_n = bleached + unbleached,
postbleaching_n = unbleached,
percent_mortality = round((bleached / prebleaching_n) * 100, 1)
) |> select(-bleached, -unbleached)
reactable::reactable(
head(colony_output, 20),
defaultPageSize = 20,
pagination = FALSE,
searchable = FALSE,
highlight = TRUE,
bordered = TRUE,
striped = TRUE
) Reproductive output estimates
Code
population_output <- reproductive_populations_postbleaching |>
as.data.frame() |>
group_by(status, species) |>
summarise(
mean_output = mean(output),
total_output = sum(output),
.groups = "drop"
) |>
pivot_wider(
names_from = status,
values_from = c(mean_output, total_output),
names_glue = "{.value}_{status}"
) |>
mutate(
mean_output_change = round(100 * (mean_output_unbleached - mean_output_bleached) / mean_output_unbleached, 1),
total_output_change = round(100 * (total_output_unbleached - total_output_bleached) / total_output_unbleached, 1),
mean_output_postbleaching = scales::comma(round(mean_output_unbleached, 1)),
mean_output_prebleaching = scales::comma(round(mean_output_bleached, 1)),
total_output_postbleaching = scales::comma(total_output_bleached),
total_output_prebleaching = scales::comma(total_output_unbleached)
) |>
select(species, mean_output_prebleaching, mean_output_postbleaching, mean_output_change,
total_output_prebleaching, total_output_postbleaching, total_output_change)
reactable::reactable(
head(population_output, 20),
defaultPageSize = 20,
pagination = FALSE,
searchable = FALSE,
highlight = TRUE,
bordered = TRUE,
striped = TRUE
)