---
title: "Publications"
author: "George Roff"
date: "2025-03-18"
format:
html:
toc: FALSE
code-fold: true
code-tools: true
execute:
echo: true
warning: false
message: false
editor: visual
---
```{r}
#| message: false
#| warning: false
#| cache: false
#| fig-height: 8
#| fig-width: 12
library(ggplot2)
library(plotly)
library(scales)
library(RColorBrewer)
conflicts_prefer(plotly::layout)
library(tidyverse)
library(scholar)
library(DT)
library(magrittr)
scholar_id <- "g_m1R7IAAAAJ"
scholar_output <- scholar::get_publications(id = scholar_id) %>%
rowwise() %>%
mutate(
author = case_when(
str_ends(author, "\\.\\.\\.") ~ get_complete_authors(id = scholar_id, pubid = pubid),
TRUE ~ author
)
) %>%
ungroup()
profile <- scholar::get_profile(scholar_id)
citation_history <- scholar::get_citation_history(id = scholar_id)
npapers <- scholar_output$title |> unique() |> length()
ncites <- scholar_output$cites |> sum()
hindex <- profile$h_index
i10 <- profile$i10_index
affiliation <- profile$affiliation
datatable(
data.frame(
a = c("Author", "Affiliation", "Total Citations", "i-10 index", "h-index", "Publications to date"),
b = c("George Roff", affiliation, ncites, hindex, i10, npapers)
) |> t(),
colnames = NULL,
options = list(
dom = 't', # show only the table body
paging = FALSE,
ordering = FALSE
),
rownames = FALSE
) %>%
formatStyle(columns = c(1, 2, 3, 4, 5, 6, 7), fontSize = '80%')
```
All publication data generated dynamically `R` (`scholar`, `wordcloud`) and `Plotly`:
### Summary statistics
```{r}
#| message: false
#| warning: false
#| cache: false
#| fig-height: 12
#| fig-width: 6
library(ggplot2)
library(plotly)
library(scales)
library(RColorBrewer)
conflicts_prefer(plotly::layout)
pubs_per_year <- scholar_output %>%
group_by(year) %>%
summarise(publications = n(), .groups = 'drop')
scholar_output_prepped <- scholar_output %>%
ungroup() %>%
arrange(desc(year), cites) %>%
mutate(
n = seq(1:nrow(.)),
author = gsub("(G Roff)", "<b>\\1</b>", author, ignore.case = TRUE), # Use HTML for bold formatting
cites = as.numeric(cites) # Ensure cites is numeric
) %>%
select(n, author, year, title, journal, number, cites) %>%
rename(
"Year" = "year",
"Authors" = "author",
"Journal" = "journal",
"Issue/Number" = "number",
"Citations" = "cites"
)
###### Plot 1 – Publications per Year
pubs_per_year <- scholar_output %>%
group_by(year) %>%
summarise(publications = n(), .groups = 'drop')
# Generate Spectral palette for publications
num_years <- nrow(pubs_per_year)
colors1 <- colorRampPalette(brewer.pal(11, "Spectral"))(num_years)
pubs_plot <- plot_ly(
pubs_per_year,
x = ~year,
y = ~publications,
type = 'bar',
marker = list(
color = colors1,
line = list(color = 'black', width = 1) # Add thin black border
),
hoverinfo = 'text',
text = ~paste0("Year: ", year, "<br>Publications: ", publications),
textfont = list(size = 6)
) %>%
layout(
title = "Publications per Year",
xaxis = list(title = "Year"),
yaxis = list(title = "Number of Publications"),
showlegend = FALSE
)
####### Plot 2 – Cumulative Citations per Year
cumulative_citations <- citation_history %>%
arrange(year) %>%
mutate(cumulative_cites = cumsum(cites))
# Step 5: Plot 3 – Citations by Publication Rank
scholar_output_ranked <- scholar_output %>%
arrange(desc(cites)) %>%
mutate(n = row_number())
# Generate Spectral palette for publication rank plot
num_ranked <- nrow(scholar_output_ranked)
colors_ranked <- colorRampPalette(brewer.pal(8, "RdYlBu"))(num_ranked)
rank_plot <- plot_ly(
scholar_output_ranked,
x = ~n,
y = ~cites,
type = 'bar',
marker = list(
color = colors_ranked,
line = list(color = 'black', width = 1) # Add thin black border
),
hoverinfo = 'text', # Only display hover text
text = ~paste0("Title: ", title, "<br>Year: ", year, "<br>Citations: ", cites),
textfont = list(size = 0) # No bar labels displayed
) %>%
layout(
title = NULL,
xaxis = list(title = "Publication Rank"),
yaxis = list(title = "Cumulative Citations"),
showlegend = FALSE,
shapes = list(
list(
type = 'line',
x0 = 0,
x1 = 1,
xref = 'paper',
y0 = 10,
y1 = 10,
line = list(
color = 'black',
width = 2,
dash = 'dot'
)
)
)
)
###### Plot 3 Cumulative citations
# Generate Spectral palette for cumulative citations
num_years2 <- nrow(cumulative_citations)
colors2 <- colorRampPalette((brewer.pal(11, "RdBu")))(num_years2)
citations_plot <- plot_ly(
cumulative_citations,
x = ~year,
y = ~cumulative_cites,
type = 'scatter',
mode = 'lines+markers',
line = list(color = 'black', width = 1), # Narrow black line
marker = list(
color = colors2,
size = 9,
line = list(color = 'black', width = 1) # Add thin black border
),
hoverinfo = 'text',
text = ~paste0("Year: ", year, "<br>Cumulative Citations: ", cumulative_cites)
) %>%
layout(
title = "",
xaxis = list(title = "Year"),
yaxis = list(title = "Cumulative Citations"),
showlegend = FALSE
)
#
# # Step 6: Display all three plots in a multi-panel layout
# subplot(
# pubs_plot, rank_plot, citations_plot,
# nrows = 1,
# titleX = TRUE,
# titleY = TRUE,
# widths = c(0.3, 0.3, 0.3), # Adjust plot widths if necessary
# margin = 0.04 # Set spacing between each plot
# ) %>%
# layout(
# xaxis = list(title = "Year / Rank", domain = c(0, 0.3), titlefont = list(size = 9), tickfont = list(size = 9)),
# xaxis2 = list(title = "Rank", domain = c(0.35, 0.65), titlefont = list(size = 9), tickfont = list(size = 9)),
# xaxis3 = list(title = "Citations", domain = c(0.7, 1), titlefont = list(size = 9), tickfont = list(size = 9)),
# yaxis = list(titlefont = list(size = 9), tickfont = list(size = 9)),
# yaxis2 = list(titlefont = list(size = 9), tickfont = list(size = 9)),
# yaxis3 = list(titlefont = list(size = 9), tickfont = list(size = 9)),
# #uniformtext = list(minsize = 6, mode = 'show'),
# margin = list(l = 50, r = 50, t = 50, b = 50, pad = 5)
# )
subplot(
pubs_plot, citations_plot, rank_plot,
nrows = 3,
titleX = TRUE,
titleY = TRUE,
margin = 0.04
) %>%
layout(
xaxis = list(title = "Year / Rank", titlefont = list(size = 9), tickfont = list(size = 9)),
xaxis2 = list(title = "Rank", titlefont = list(size = 9), tickfont = list(size = 9)),
xaxis3 = list(title = "Citations", titlefont = list(size = 9), tickfont = list(size = 9)),
yaxis = list(titlefont = list(size = 9), tickfont = list(size = 9)),
yaxis2 = list(titlefont = list(size = 9), tickfont = list(size = 9)),
yaxis3 = list(titlefont = list(size = 9), tickfont = list(size = 9)),
margin = list(l = 50, r = 50, t = 50, b = 50, pad = 5)
)
```
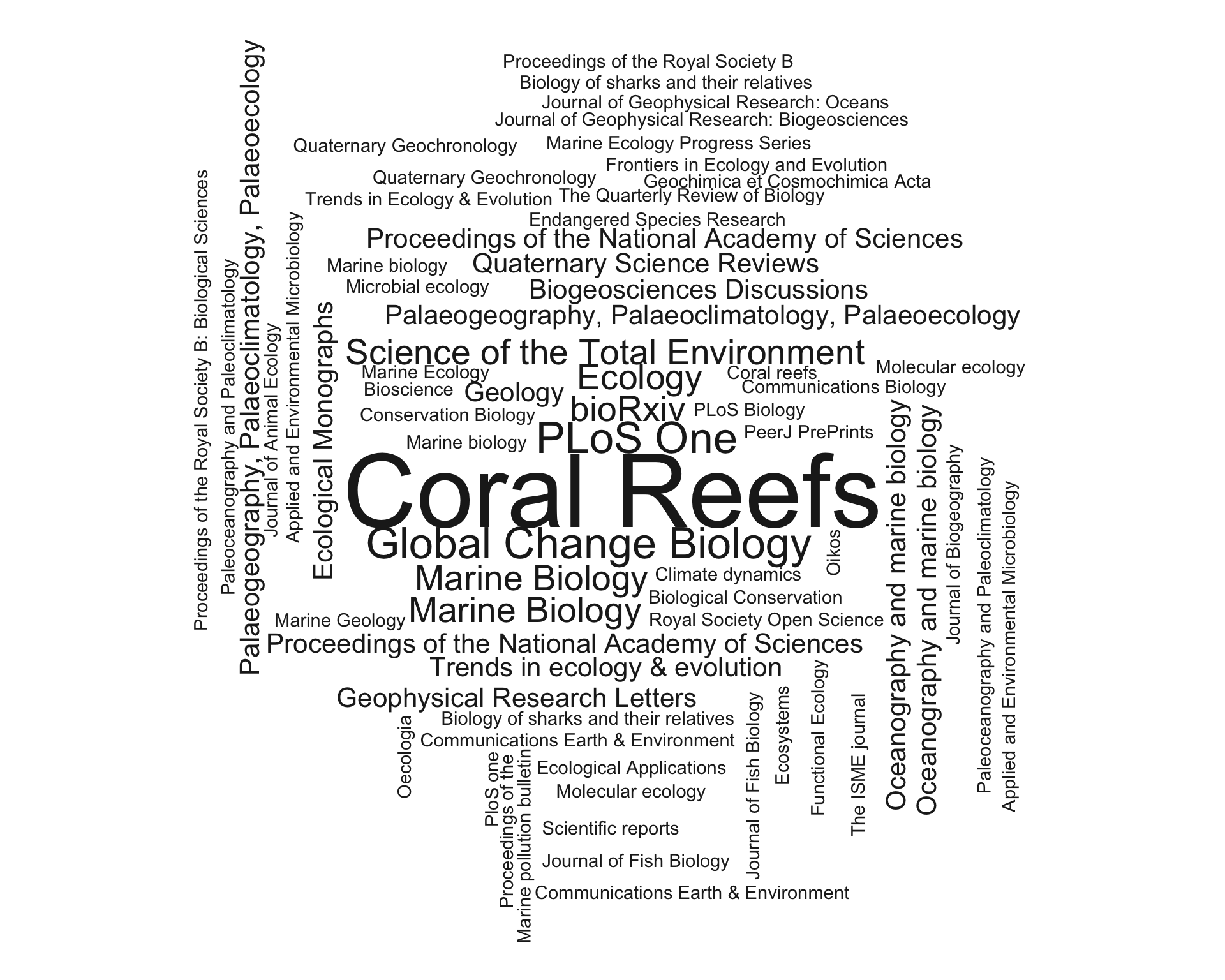
### Publication journals
```{r}
#| message: false
#| warning: false
#| fig-height: 8
#| fig-width: 10
library(wordcloud)
library(grid)
library(cowplot)
library(gridGraphics)
library(png)
library(gtable)
# Define keyword lookup
subject_keywords <- list(
Ecology = c("ecology", "oikos", "oecologia", "ecological", "Ecosystems"),
MarineBiology = c("coral reefs", "marine biology", "PLoS ONE", "proceedings", "Scientific", "Fish", "total", "Oceanography", "sharks"),
Geology = c("palaeo", "geology", "earth", "Quaternary", "Paleoceanography"),
Microbiology = c("microbiology", "Molecular", "isme"),
Climate = c("National", "Open", "Climate", "bioRxiv"),
Geochemistry = c("geochem", "Geophysical", "Geochronology", "Geochimica", "Biogeosciences"),
Conservation = c("pollution", "peer", "global", "conservation","Communications", "endangered", "Biology"),
Evolution = c("bioscience", "Biogeography")
)
# Assign subject(s)
journal_subjects <- scholar_output_prepped %>%
mutate(
subject = sapply(Journal, function(j) {
matches <- names(subject_keywords)[
sapply(subject_keywords, function(keywords) {
any(grepl(paste(keywords, collapse = "|"), j, ignore.case = TRUE))
})
]
paste(matches, collapse = ";")
})
)
# Count frequency of each journal per subject
subject_freq <- journal_subjects %>%
filter(subject != "") %>%
separate_rows(subject, sep = ";") %>%
group_by(subject, Journal) %>%
summarise(n = n(), .groups = "drop") %>%
arrange(subject, desc(n)) %>%
mutate(freq_scaled = log1p(n))
set.seed(1)
wordcloud(
words = subject_freq$Journal,
freq = subject_freq$n,
min.freq = 1,
max.words = Inf,
color="#202020",
# colors = brewer.pal(4, "Spectral"),
scale = c(5,0.5),
random.order = FALSE,
use.r.layout = TRUE,
rot.per=0.3
)
set.seed(NULL)
```
# Selected publications
```{r}
#| message: false
#| warning: false
#| cache: false
#| fig-height: 8
#| fig-width: 12
## All publications to bib
# Convert to BibTeX format
scholar_bib <- scholar_output %>%
mutate(
bibtex_entry = paste0(
"@article{", pubid, ",\n",
" title = {", title, "},\n",
" author = {", author, "},\n",
" journal = {", journal, "},\n",
" year = {", year, "},\n",
" volume = {", sub(" \\(.*", "", number), "},\n", # Extract volume before '('
" number = {", sub(".*\\((.*)\\).*", "\\1", number), "},\n", # Extract issue inside '()'
" pages = {", cites, "},\n",
" cid = {", cid, "},\n",
" pubid = {", pubid, "}\n",
"}"
)
) %>%
pull(bibtex_entry)
#writeLines(scholar_bib, "scholar_output.bib")
## select publications to bib
search_terms <- c("Global disparity", "sharks on coral reefs", "evolutionary history", "seascapes", "decline of branching", "decoupled in Holocene")
scholar_bib <- scholar_output %>%
filter(sapply(title, function(x) any(grepl(paste(search_terms, collapse = "|"), x, ignore.case = TRUE)))) |>
mutate(
bibtex_entry = paste0(
"@article{", pubid, ",\n",
" title = {", title, "},\n",
" author = {", author, "},\n",
" journal = {", journal, "},\n",
" year = {", year, "},\n",
" volume = {", sub(" \\(.*", "", number), "},\n", # Extract volume before '('
" number = {", sub(".*\\((.*)\\).*", "\\1", number), "},\n", # Extract issue inside '()'
" pages = {", cites, "},\n",
" cid = {", cid, "},\n",
" pubid = {", pubid, "}\n",
"}"
)
) %>%
pull(bibtex_entry)
writeLines(scholar_bib, "select_scholar_output.bib")
# Function to lighten a vector of colors
lighten_colors <- function(colors, factor = 0.5) {
if (factor < 0 || factor > 1) stop("Factor must be between 0 and 1")
col2rgb(colors) %>%
t() %>%
apply(1, function(col) {
rgb(
red = col[1] + (255 - col[1]) * factor,
green = col[2] + (255 - col[2]) * factor,
blue = col[3] + (255 - col[3]) * factor,
maxColorValue = 255
)
})
}
pubs_per_year <- scholar_output %>%
group_by(year) %>%
summarise(publications = n(), .groups = 'drop')
scholar_output_prepped <- scholar_output %>%
ungroup() %>%
arrange(desc(year), cites) %>%
mutate(
n = seq(1:nrow(.)),
author = gsub("(G Roff)", "<b>\\1</b>", author, ignore.case = TRUE), # Use HTML for bold formatting
cites = as.numeric(cites) # Ensure cites is numeric
) %>%
select(n, author, year, title, journal, number, cites) %>%
rename(
"Year" = "year",
"Authors" = "author",
"Journal" = "journal",
"Issue/Number" = "number",
"Citations" = "cites"
)
# Generate colors using RColorBrewer with adjusted alpha
num_colors <- nrow(scholar_output_prepped)
color_palette <- colorRampPalette(rev(RColorBrewer::brewer.pal(9, "RdYlBu")))(num_colors)
color_palette <- lighten_colors(color_palette, factor = 0.4) # Set alpha to 0.7
# Map colors to the Citations column
scholar_output_prepped <- scholar_output_prepped %>%
filter(grepl("Palaeoecological evidence", title) |
grepl("Global disparity", title) |
grepl("Decadal‐scale rates of reef erosion", title) |
grepl("Seascape", title) |
grepl("Rapid accretion of inshore reef", title) |
grepl("Reassessing Shark-Driven Trophic", title) |
grepl("Decline of coastal apex shark populations", title) |
grepl("Reef accretion and coral growth rates", title) |
grepl("Evolutionary history drives", title) |
grepl("Revisiting the paradigm of shark‐driven", title) |
grepl("Climate change impacts on mesophotic regions", title) |
grepl("Reef accretion and coral growth rates", title) |
grepl("Cryptic coral recruits as dormant “seed banks”", title)
) |>
mutate(
cite_color = color_palette[rank(Citations, ties.method = "first")]
)
datatable(
scholar_output_prepped %>% select(-cite_color) %>% mutate(n=seq(1:n())),
escape = FALSE,
options = list(
search = list(regex = TRUE, caseInsensitive = FALSE),
autoWidth = FALSE,
dom = 't<"bottom"ip>',
pageLength = 100,
columnDefs = list(
list(className = 'dt-center', targets = "_all"),
list(width = '10%', targets = 0),
list(width = '30%', targets = 1),
list(width = '10%', targets = 2),
list(width = '30%', targets = 3),
list(width = '10%', targets = 4),
list(width = '10%', targets = 5),
list(width = '10%', targets = 6)
)
),
rownames = FALSE,
class = "cell-border stripe"
) %>%
formatStyle( 'Citations',
backgroundColor = styleEqual(scholar_output_prepped$Citations, scholar_output_prepped$cite_color),
color = "black", fontSize = '60%'
) %>%
formatStyle(columns = c(1, 2, 3, 4, 5, 6, 7), fontSize = '80%')
```